3D DeepLabCut#
In this repo we directly support 2-camera based 3D pose estimation. If you want n camera support, plus nicer optimization methods, please see our work that was published at ICRA 2021 on strong baseline 3D models (and a 3D dataset). In the link you will find how we optimize 6+ camera DLC output data for cheetahs (and see more below).

ATTENTION: Our code base in this repo assumes you:#
A. You have 2D videos and a DeepLabCut network to analyze them as described in the main documentation. This can be with multiple separate networks for each camera (less recommended), or one network trained on all views - recommended! (See Nath*, Mathis* et al., 2019). We also support multi-animal 3D with this code (please see Lauer et al. 2022).
B. You are using 2 cameras, in a stereo configuration, for 3D*.
C. You have calibration images taken (see details below!).
*If you need more than 2 camera support:#
Here are other excellent options for you to use that extend DeepLabCut:
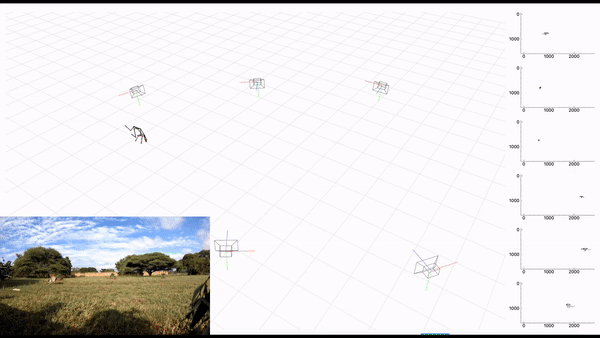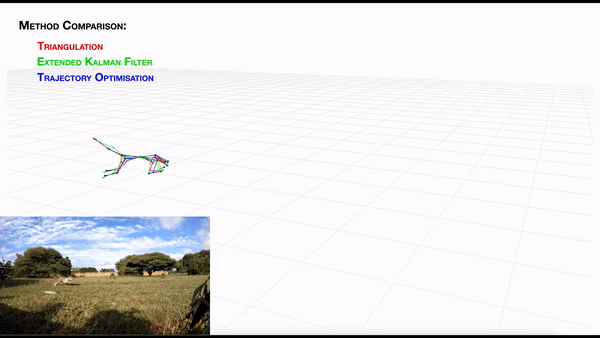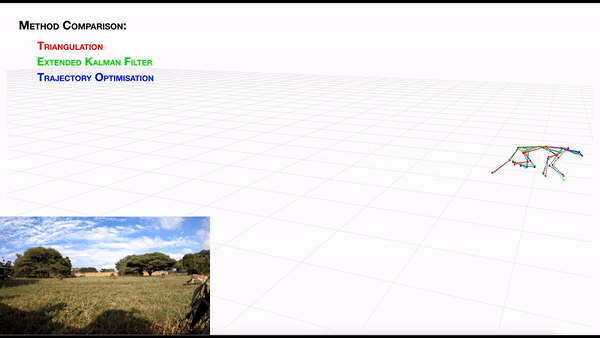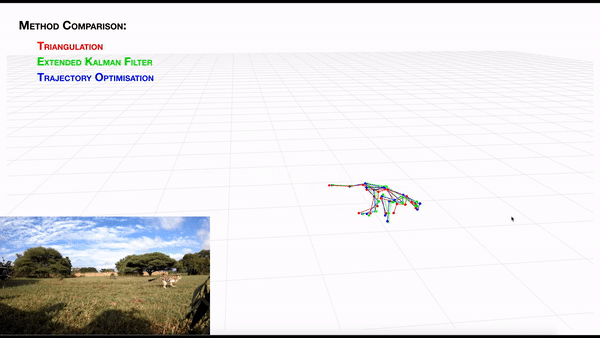
AcinoSet; n-camera support with triangulation, extended Kalman filtering, and trajectory optimization code (see video to the right for a min demo, courtesy of Prof. Patel), plus a GUI to visualize 3D data. It is built to work directly with DeepLabCut (but currently tailored to cheetah’s, thus some coding skills are required at this time).
anipose.org; a wrapper for 3D deeplabcut that provides >3 camera support and is built to work directly with DeepLabCut. You can
pip install aniposeinto your DLC conda environment.Argus, easywand or DLTdv w/DeepLabCut see backyardbiomech/DLCconverterDLT; this can be used with the the highly popular Argus or DLTdv tools for wand calibration. As of Summer, 2025, Argus now supports direct import and export of DeepLabCut output files in the GUI with new workflow documentation
Jump in with direct DeepLabCut 2-camera support:#
single animal DeepLabCut and multi-animal DeepLabCut (maDLC) projects are supported:


(1) Create a New 3D Project:#
Watch a DEMO VIDEO on how to use this code, and check out the Notebook here!
You will run this function one time per project; a project is defined as a given set of cameras and calibration images. You can always analyze new videos within this project.
The function create_new_project_3d creates a new project directory specifically for converting the 2D pose to 3D pose, required subdirectories, and a basic 3D project configuration file. Each project is identified by the name of the project (e.g. Task1), name of the experimenter (e.g. YourName), as well as the date at creation.
Thus, this function requires the user to enter the name of the project, the name of the experimenter and number of cameras to be used. Currently, DeepLabCut supports triangulation using 2 cameras, but will expand to more than 2 cameras in a future version.
To start a 3D project type the following in ipython:
deeplabcut.create_new_project_3d("ProjectName", "NameofLabeler", num_cameras=2)
TIP 1: you can also pass working_directory="Full path of the working directory" if you want to place this folder
somewhere beside the current directory you are working in. If the optional argument working_directory is unspecified,
the project directory is created in the current working directory.
TIP 2: you can also place config_path3d in front of deeplabcut.create_new_project_3d to create a variable that holds
the path to the config.yaml file, i.e. config_path3d=deeplabcut.create_new_project_3d(... Or, set this variable for
easy use. Please note that config_path3d='Full path of the 3D project configuration file'.
This function will create a project directory with the name Name of the project+name of the experimenter+date of creation of the project+3d in the Working directory. The project directory will have subdirectories: calibration_images, camera_matrix, corners, and undistortion. All the outputs generated during the course of a project will be stored in one of these subdirectories, thus allowing each project to be curated in separation from other projects.
The purpose of the subdirectories is as follows:
calibration_images: This directory will contain a set of calibration images acquired from the two cameras. A calibration image can be acquired using a printed checkerboard and its pair wise images are taken from both the cameras to consider as a set of calibration images.
camera_matrix: This directory will store the parameter for both the cameras as a pickle file. Specifically, these pickle files contain the intrinsic and extrinsic camera parameters. While the intrinsic parameters represent a transformation from 3-D camera’s coordinates into the image coordinates, the extrinsic parameters represent a rigid transformation from world coordinate system to the 3-D camera’s coordinate system.
corners: As a part of camera calibration, the checkerboard pattern is detected in the calibration images and these patterns will be stored in this directory. Each row of the checkerboard grid is marked with a unique color.
undistortion: In order to check for calibration, the calibration images and the corresponding corner points are undistorted. These undistorted images are overlaid with undistorted points and will be stored in this directory.
Here is an overview of the calibration and triangulation workflow that follows:

(2) Take and Process Camera Calibration Images:#
(CRITICAL!) You must take images of a checkerboard to calibrate your images. Here are example boards you could print and use (mount it on a flat, hard surface!): https://markhedleyjones.com/projects/calibration-checkerboard-collection.
You must save the image pairs as .jpg files.
They should be named with the camera-# as the prefix, i.e. camera-1-01.jpg and camera-2-01.jpg for the first pair of images. Please note, this cannot be changed after the project is created.
TIP: If you want to take a short video (vs. snapping pairs of frames) while you move the checkerboard around, you
can use this command inside your conda environment (but outside of ipython!) to convert the video to .jpg frames
(this will take the first 20 frames (set with -vframes) and name them camera-1-001.jpg, etc; edit appropriately):
ffmpeg -i videoname.mp4 -vframes 20 camera-1-%03d.jpg
While taking the images:
Keep the orientation of the checkerboard the same and do not rotate it more than 30 degrees. Rotating the checkerboard circular will change the origin across the frames and may result in incorrect order of detected corners.
Cover several distances, and within each distance, cover all parts of the image view (all corners and center).
Use a checkerboard as big as possible, ideally with at least 8x6 squares.
Aim for taking at least 30-70 pair of images, as after corner detection, some of the images might need to be discarded due to either incorrect corner detection or incorrect order of detected corners.
You can take the images as a series of .jpg images, or a video where you post-hoc pair sync’d frames (see tip above).
The camera calibration is an iterative process, where the user needs to select a set of calibration images where the
grid pattern is correctly detected. The function deeplabcut.calibrate_cameras(config_path)
extracts the grid pattern from the calibration images and store them under the corners directory. The grid pattern
could be 8x8 or 5x5 etc. We use a pattern of the 8x6 grid to find the internal corners of the checkerboard.
In some cases, it may happen that the corners are not detected correctly or the order of corners detected in the camera-1 image and camera-2 image is incorrect. You need to remove these pair of images from the calibration_images folder as they will reduce the calibration accuracy.
To begin, please place your images into the calibration_images directory.
(CRITICAL!) Edit the config.yaml file to set the camera names; note that once this is set, do not change the names!
Then, run:
deeplabcut.calibrate_cameras(config_path3d, cbrow=8, cbcol=6, calibrate=False, alpha=0.9)
NOTE: you need to specify how many rows (cbrow) and columns (cbcol) your checkerboard has (beware, we count
edges between squares and not squares themselves, so for a 8 x 8 squares checkerboard set cbrow=7 and cbcol=7).
Also, first set the variable calibrate to False, so you can remove any faulty images. You need to visually
inspect the output to check for the detected corners and select those pair of images where the corners are correctly
detected. Please note, If the scaling parameter alpha=0, it returns undistorted image with minimum unwanted pixels.
So it may even remove some pixels at image corners. If alpha=1, all pixels are retained with some extra black images.
Here is what they might look like:


Once all the set of images has been selected (namely, delete from the folder any bad pairs!) where the corners and their orders are detected correctly, then the two cameras can be calibrated using:
deeplabcut.calibrate_cameras(config_path3d, cbrow=8, cbcol=6, calibrate=True, alpha=0.9)
This computes the intrinsic and extrinsic parameters for each camera. A re-projection error is also computed using the
intrinsic and extrinsic parameters which provide an estimate of how good the parameters are. The transformation between
the two cameras is estimated and the cameras are stereo calibrated. Furthermore, the above function brings both the
camera image plane to the same plane by computing the stereo rectification. These parameters are stored as a pickle file
named as stereo_params.pickle under the directory camera_matrix.
Once you have run this for the project, you do not need to do so again (unless you want to re-calibrate your cameras); be advised, if you do re-calibrate, you may want to clearly mark which videos are analyzed with “old” vs. “new” calibration images.
(3) Check for Undistortion:#
In order to check how well the stereo calibration is, it is recommended to undistort the calibration images and the corner points using camera matrices and project these undistorted points on the undistorted images to check if they align correctly. This can be done in deeplabcut as:
deeplabcut.check_undistortion(config_path3d, cbrow=8, cbcol=6)
Each calibration image is undistorted and saved under the directory undistortion. A plot with a pair of undistorted
camera images with its undistorted corner points overlaid is also stored. Please visually inspect this image. All the
undistorted corner points from all the calibration images are triangulated and plotted for the user to visualize for any
undistortion related errors. If they are not correct, go check and revise the calibration images (then repeat the
calibration and this step)!
(4) Triangulation –> Take your 2D to 3D!#
If there are no errors in the undistortion, then the pose from the 2 cameras can be triangulated to get the 3D DeepLabCut coordinates!
(CRITICAL!) Name the video files in such a way that the file name contains the name of the cameras as specified
in the config file. e.g. if the cameras as named as camera-1 and camera-2 (or cam-1, cam-2 etc.) then the
video filename must contain this naming, i.e. this could be named as rig-1-mouse-day1-camera-1.avi and
rig-1-mouse-day1-camera-2.avi or could be rig-1-mouse-day1-camera-1-date.avi and
rig-1-mouse-day1-camera-2-date.avi.
Note that to correctly pair the videos, the file names otherwise need to be the same!
If helpful, here is the software we use to record videos.
(CRITICAL!) You must also edit the 3D project config.yaml file to denote which DeepLabCut projects have the information for the 2D views.
Of critical importance is that you need to input the same body part names as in the config.yaml file of the 2D project.
You must set the snapshot to use inside the 2D config file (default is -1, namely the last training snapshot of the network).
You need to set a “scorer 3D” name; this will point to the project file and be set in future 3D output file names.
You should define a “skeleton” here as well (note, this is not rigid, it just connects the points in the plotting step). Not every point needs to be “skeletonized”, i.e. these points can be a subset of the full body parts list. The other points will just be plotted into the 3D space. Here is how the config.yaml looks with some example inputs:

(CRITICAL!) This step will also run the equivalent of analyze_videos (in 2D) for you and then apply a median
filter to the 2D data (filterpredictions=True)! If you already ran the 2D analysis and there is a filtered output
file, it will take this by default (otherwise it will take your unfiltered 2D analysis files)!
Next, pass the config_path3d and now the video folder path, which is the path to the folder where all the videos
from two cameras are stored. The triangulation can be done in deeplabcut by typing:
deeplabcut.triangulate(
config_path3d,
"/yourcomputer/fullpath/videofolder",
filterpredictions=True/False
)
NOTE: Windows users, you must input paths as: r`C:\Users\computername\videofolder' or
C:\\Users\\computername\\videofolder'.
TIP: Here are all the parameters you can pass:
Parameters
----------
config : string
Full path of the config.yaml file as a string.
video_path : string
Full path of the directory where videos are saved.
videotype: string, optional
Checks for the extension of the video in case the input to the video is a directory.
Only videos with this extension are analyzed. The default is ``.avi``
filterpredictions: Bool, optional
Filter the predictions by fitting median (by default) or arima filter. If specified it should be either ``True`` or ``False``.
filtertype: string
Select which filter, 'arima' or 'median' filter.
gputouse: int, optional. Natural number indicating the number of your GPU (see number in nvidia-smi). If you do not have a GPU put None.
See: https://nvidia.custhelp.com/app/answers/detail/a_id/3751/~/useful-nvidia-smi-queries
destfolder: string, optional
Specifies the destination folder for analysis data (default is the path of the video)
save_as_csv: bool, optional
Saves the predictions in a .csv file. The default is ``False``; if provided it must be either ``True`` or ``False``
track_method: str, optional
Method used for tracking: "box" or "ellipse"
The triangulated file is now saved under the same directory where the video files reside (or the destination folder
you set)! This can be used for future analysis. This step can be run at anytime as you collect new videos, and easily
added to your automated analysis pipeline, i.e. such as replacing
deeplabcut.triangulate(config_path3d, video_path) with deeplabcut.analyze_videos (as if it’s not analyzed in 2D
already, this function will take care of it ;):

(5) Visualize your 3D DeepLabCut Videos:#
In order to visualize both the 2D videos with tracked points plut the pose in 3D, the user can create a 3D video for
certain frames (these are large files, so we advise just looking at a subset of frames). The user can specify the config
file, the path of the triangulated file folder, and specify the start and end frame indices to create a 3D labeled
video. Note that the triangulated_file_folder is where the newly created file that ends with
yourDLC_3D_scorername.h5 is located. This can be done using:
deeplabcut.create_labeled_video_3d(
config_path,
["triangulated_file_folder"],
start=50,
end=250
)
TIP: (see more parameters below) You can set how the axis of the 3D plot on the far right looks by changing the
variables xlim, ylim, zlim and view. Your checkerboard_3d.png image which was created above will show you the
axis ranges. Here is an example:

View is used to set the elevation and azimuth of the axes (defaults are [113, 270], and you should play around to find
the view-point you like!). Also note that the video is created from a set of .png files in a “temp” directory, so as
soon as you run this command you can open the first image, and if you don’t like the view, hit CNTRL+C to stop, edit
the values, and start again!
Other optional parameters include: here
videofolder: string
Full path of the folder where the videos are stored. Use this if the videos are stored in a different location other than where the triangulation files are stored. By default is ``None`` and therefore looks for video files in the directory where the triangulation file is stored.
trailpoints: int
Number of previous frames whose body parts are plotted in a frame (for displaying history). Default is set to 0.
videotype: string
Checks for the extension of the video in case the input is a directory.
Only videos with this extension are analyzed. The default is ``.avi``
view: list
A list that sets the elevation angle in z plane and azimuthal angle in x,y plane of 3d view. Useful for rotating the axis for 3d view
xlim: list
A list of integers specifying the limits for xaxis of 3d view. By default it is set to [None,None], where the x limit is set by taking the minimum and maximum value of the x coordinates for all the bodyparts.
ylim: list
A list of integers specifying the limits for yaxis of 3d view. By default it is set to [None,None], where the y limit is set by taking the minimum and maximum value of the y coordinates for all the bodyparts.
zlim: list
A list of integers specifying the limits for zaxis of 3d view. By default it is set to [None,None], where the z limit is set by taking the minimum and maximum value of the z coordinates for all the bodyparts.
draw_skeleton: bool
If True adds a line connecting the body parts making a skeleton on on each frame. The body parts to be connected and the color of these connecting lines are specified in the config file. By default: True
color_by : string, optional (default='bodypart')
Coloring rule. By default, each bodypart is colored differently.
If set to 'individual', points belonging to a single individual are colored the same.
figsize: tuple[int, int], optional, default=(80, 8)
Size of the figure
fps: int, optional, default=30
Frames per second
dpi: int, optional, default=300
Dots per inch (resplution)
If you use this code:#
We kindly ask that you cite Mathis et al, 2018 & Nath*, Mathis*, et al., 2019. If you use 3D multi-animal: Lauer et al. 2022.